14 July 2020
Dr Cate Williams: IBERS, Aberystwyth University.
- Genomic testing of dairy bulls has revolutionised sire selection – offering large amounts of accurate and reliable information enabling an informed choice.
- Testing female animals – particularly heifers – could offer similar advantages, like rapid genetic gain and accurate selection of replacements which may result in more profitable animals.
- Genomic testing is becoming cheaper and more accessible and provides more reliable information than a daughter-based evaluation, but it is still early days and case studies are not yet available.
- Genes for traits of interest have differing levels of heritability and will interact with one another, so it is important to consider this when making selections.
The recent, strategic use of genomic testing in young dairy bulls has revolutionised the way dairy farmers select sires. Where previously, a bull must be daughter proven, which may take up to 6 years, young bulls can now be genetically tested as calves (<1 year old), which alongside dam and sire information gives a detailed report of that animal’s genetic potential without the need for progeny. This has accelerated the rate of genetic gain, reduced the generation interval and hugely improved accuracy of selection in dairy herds in developed countries around the world. However, this represents just one side of the coin, what about the cow or heifer? Breeding a bull with “excellent” or “very good” genetic potential to a cow with poor or only fair genetic potential means that it’s unlikely that the offspring will ever reach similar potentials to the sire. In the long term, the use of genomic testing on cows and heifers could offer similar benefits to testing bulls, enabling highly accurate selection of replacements. Selecting for only the best replacement heifers and those that best suit the farming system can have a positive effect on farm profitability.
How does it work?
Genetic testing is an umbrella term for any test that analyses changes in the DNA sequence or chromosome structure, it may target DNA regions or a single gene. Genomic testing, on the other hand, refers to the analysis of the whole genome (that is all of the DNA of an organism) enabled by new, rapid, high throughput sequencing technologies. Fully sequenced in 2009, the bovine genome comprises 22,000 individual genes so the need for advanced technology – both hardware and software – becomes evident. The test requires a sample from the animal, usually in the form of tissue or hair accompanied by as much pedigree information as is available (from both the sire and dam’s sides). DNA is extracted from the sample, prepared and then sequenced, this sequencing data is then transferred onto a computer where it must be assembled in the correct order and then interpreted – the average size of the bovine genome is the same size as a human one at approx. 3 Gb (gigabases) or 3 billion base pairs. The sequence data should now be organised into genes and chromosomes and so a picture of the animal’s genetic makeup begins to emerge. Key to any kind of genetic sequencing is the construction of a reference population. To organise the sample data into genes and chromosomes, it is compared to other known, correct sequences, a bit like using the picture on the front of the box to correctly assemble a jigsaw puzzle. To this end, DNA from a wide variety of breeds and animals of different genetic merit must be collected to allow for accurate annotation of subsequent samples. To generate a large enough pool of reference data, information is shared between countries – currently, the UK shares genomic information for Holstein, Guernsey, Jersey and Ayrshire cattle with much of Europe, Canada and the USA.
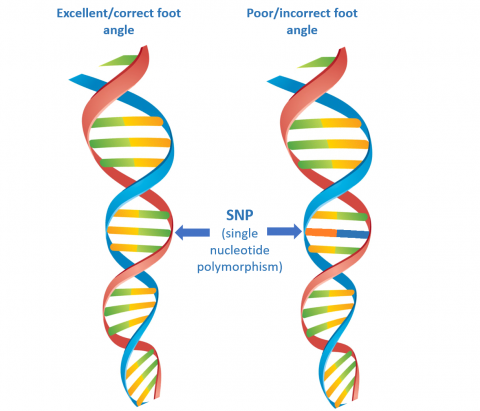
To draw comparisons between the reference population and the target animal, markers (single nucleotide polymorphisms or SNPs) for traits of interest are identified and compared between the sample and the reference population (Figure 1). For example, are the SNPs in our target animal that are responsible for foot angle similar to those with very good foot angles in the reference population, or are they more comparable to those with a poor foot angle (Figure 1)? This systematic comparison spans the entire genome and addresses a plethora of traits (or indices) which come together to give Genomic Estimated Breeding Values (GEBVs) - an incredibly detailed picture of the animal’s genetic potential.
Figure 1: Basic, diagrammatic representation of a hypothetical single nucleotide polymorphism (SNP) responsible for foot angle.
Pros and cons
The benefits of genomic selection are numerous and far-reaching. The identification of cows with the highest genetic potential accelerates genetic gain by facilitating accurate identification of cull, sale or replacement animals. Retaining only those with good genetic potential can have a range of knock-on effects on fertility, calving, milk production and quality, animal health, feed costs, longevity and overall profitability, to name a few. This information is particularly useful when used more than once in the animal’s lifetime, i.e. in replacement heifers when the decision is made to retain them and also which bull to put them to. The most benefit is seen in traits with low heritability rates and those that can only be measured later in life after the farmer has already committed a great deal of time and money to the animal. Examples include lifespan temperament and fertility – traits that may only become apparent after several years in the herd.
Over the past 20 years, genomic selection has become more and more accurate as technology has developed and as more information has been gathered. The accuracy of testing depends on several factors: the size of the reference population, the heritability of the trait in question and the number of SNPs relating to the trait. For example, comparing 20 Holstein heifers from Wales to 1,000 Jersey cows from Canada for a trait with very few markers is not going to give very accurate information. On the other hand, comparing those same 20 Welsh Holsteins to records from 100,000 other Holsteins from the UK, Canada and the USA for a trait with 100 related SNPs will likely give very accurate results. At present, the reliability of GEBVs for production traits is often 70% or greater, which is twice that of traditional parent averages computed from pedigrees. There is a large range of traits that can be tested which cover reproduction, health/longevity and productivity.
A key example of this is the Economic Breeding Index (EBI) developed in Ireland, designed to help farmers select the most profitable bulls for breeding and heifers for herd replacements. A recent study comparing ‘elite’ and ‘national average’ Holstein Friesian cows revealed that cows performing at a national average produced a higher volume of milk than those in the elite group, however, milk from the elite group contained higher levels of milk fat and protein, resulting in similar yields of solids-corrected milk. When offered different diets (e.g. forage-based, supplemented with concentrates or high concentrate diets) these differences were maintained, with elite cows showing relatively little difference in milk volume and solids regardless of diet. The study highlights the success of the Irish EBI system in reducing milk volume whilst increasing milk solid yields. Another benefit of genomic testing is the prevention or slowing of inbreeding and eradication of genetic diseases - these reasons may be insufficient justification in isolation, but when coupled with other factors make genomic testing a more attractive prospect.
One of the key drawbacks of GEBVs is the cost, which has been a central issue for all producers since the technology became available. However, as technology has developed, it has become more and more affordable, especially when weighed against the potential for more profitable animals. One key, in-depth study modelled the potential outcomes stemming from the use of genomic testing in heifers. Results consistently suggested that genetic gains and increases in farm profitability would more than compensate for the initial cost of the test. It is also important to remember that GEBVs are reporting genetic potential, i.e. what the animal could achieve under perfect conditions. This is not a guarantee that the animal will achieve its potential as many other environmental factors will also play a role in an animal’s performance.
The impact of genomic testing
There is a body of scientific evidence to support the use of genomic testing in cows/heifers, however, such studies are based on modelling. The concept of genomic testing in females is relatively new when compared to testing bulls and as such, it will likely be a little more time before case studies become available. An example of such work may be found in the Farming Connect demonstration farm at Mountjoy, where genomic testing is being used to select their replacement heifers, with a particular interest in the TB advantage and milk solids (protein and fat) indices. The project aims to use genomic screening to select heifers that best suit their farm whilst also investigating the use of satellite grass mapping and herbal leys with the end goal of reducing the farm’s environmental footprint but improving animal performance and profitability.
Indeed, preliminary results from the European Innovation Partnership (EIP) Wales project applying genomic testing to Welsh dairy herds have been promising. The project is using genomic screening to decide which animals to breed to a beef bull (those with lower genetic merit) and which to breed to a dairy bull (those with higher genetic merit). The project focuses on the genomic Profitable Lifetime Index (PLI, £) which includes both health and production traits. Thus far, significant differences between PLI scores generated through genomic testing and those from parent averages have been identified. Without the additional information and clarity provided by genomic testing, around 76 animals would have been incorrectly categorised and put to the wrong bull. This could contribute to slower genetic gain, reduce the longevity of the heifer/cow and negatively affect future generations. The interim report highlights the common misconception that an animal sold with a genomic test has poor genetic merit. Each farm needs slightly different qualities in their stock – whilst one may wish to increase milk yield another may want to focus on disease resistance. So, a genomic test can offer a buyer an additional level of confidence when making a purchase. Throughout the project, the group will aim to collect even further data (e.g. milk yield and quality, mastitis, level, fertility data and lameness issues) to present a robust evaluation of genomic screening and it’s impact on each dairy enterprise.
As more data is collected and added to reference populations and SNPs are discovered and validated, a look at the heritability of target traits may help to give an idea of how quickly genomic testing will improve a certain trait (Table 1). A high heritability score indicates that a gene(s) is more likely to be passed on to offspring meaning that improvements in the related trait would likely emerge quicker than one with a low heritability score. For example, the TB Advantage index created by AHDB has a relatively low heritability score of ~0.1, similar to that of somatic cell counts which average 0.16 (Table 1). Traits related to feed efficiency (DMI and RFI) and milk fat and protein content, on the other hand, are moderately heritable ranging between 0.30 and 0.43 (Table 1). There is also the added complexity of interactions and linkages between genes responsible for these traits, for example, it is well established that fertility is negatively impacted when milk yield is selected for in dairy cattle. The mapping of such interactions is ongoing and will likely play an important role in selections decisions in the future.
|
Trait |
Heritability scores |
Avg. |
|||||||||||
|
Fat |
|
|
|
|
|
|
|
0.39 |
|||||
|
Protein |
|
|
|
|
|
|
|
0.41 |
|||||
|
SCCs |
|
|
|
|
|
|
|
|
|
0.16 |
|||
|
DMI |
|
|
|
|
|
|
|
|
|
0.43 |
|||
|
RFI |
|
|
|
|
|
|
|
0.30 |
|||||
Table 1: Heritability estimates of some key traits in dairy cattle. Estimates are collected from a range of studies across a variety of dairy breeds and geographical locations with the average figure from all the studies given in the right hand column. SCCs: Somatic Cell Counts; DMI: Dry Matter Intake; RFI: Residual Feed Intake.
Summary
Genomic testing of dairy bulls has revolutionised the selection of sires across the globe, resulting in younger sires, a shorter generation interval and rapid genetic gains. This in-depth genomic information is also more accurate and reliable than ‘daughter-based’ evaluation and has allowed farmers to make more informed and confident decisions when selecting sires. Additional benefits include a reduction in inbreeding and the potential for eradication of genetic diseases. A key barrier to the use of genomic testing, in general, has historically been the cost, but as technologies mature and become more efficient the cost of screening is also decreasing. When weighed against the potential for genetic gains and increases in farm profitability the initial cost of the test is recouped several times over. Genomic screening has the potential to deliver a very large amount of data, so the farmer must decide on areas for focus according to each individual farm. The most benefit is predicted to be in screening for traits with low heritability rates or that can only be measured later in life after the farmer has already committed a great deal of time and money to the animal. Whilst not the only deciding factor, heritability plays an integral role in genomic evaluations – as traits with low heritability scores will likely take longer to affect a noticeable difference in the herd. Ongoing research to refine and validate these heritability values and their interactions with one another is paramount to provide the industry with the most accurate information possible.